- The human genome is the complete set of genetic material of an individual. The exome is composed of all the protein coding exons within the genome and comprises about 2% of the human genome.
- Whole exome sequencing (WES) is a technique for sequencing all the protein-coding genes in a genome. The goal of this approach is to accurately identify genetic variants in the target regions, and to do this at a much lower cost than whole-genome sequencing (WGS). Although the exome is a small part of the genome about 85% of all known disease-causing variants are located in the exome. WES has proven to be an efficient method to determine the genetic basis of many Mendelian or single gene disorders and common polygenic diseases, as well as more complicated diseases such as cancer.
- Sequencing is the process of determining the order of nucleotides in our DNA, the nucleotides are the building blocks of our DNA and are the set of four letters that make up the genetic code. Next-generation sequencing (NGS) is a sequencing technique that can allow rapid sequencing of large amounts of DNA at the same time.
- WHOLE EXOME SEQUENCING analyses all genes associated with a phenotype based on the clinical and molecular evidence according to several reference databases (i.e.: OMIM database: https://omim.org/). A team of geneticists and specialized clinicians interpret the results by utilizing information from the latest publications and databases to produce a comprehensive clinical statement.
- Due to the technological advances in genetic testing, WES can now be considered as a first-line genetic test in complex genetic cases. WES is increasingly used in healthcare and research to identify genetic variants that cause disease and to confirm diagnoses at a molecular level. Variants in the DNA that are not located in the exons but affect gene activity and protein production can be detected using whole genome sequencing (WGS), which pans the entire genome of an individual.
WES – Whole Exome Sequencing

-
WES Diagnostic
- Whole Exome Sequencing can be used as a diagnostic tool for patients with complex genetic disorders, where the correct diagnosis is difficult to establish due to overlapping symptoms, complicated medical histories or in cases where previous genetic testing has not yielded conclusive results.
-
WES Diagnostic Index
- It is also performed on the parents of an affected individual or a couple with a family history of a known condition to identify and report any variants relating to the phenotype in question. In addition, a screening test is offered to help identify common recessive variants between couples to attempt and prevent any further genetic disorders. This WES is recommended for affected individuals who have a suspected genetic diagnosis, to help diagnose affected individuals who have multiple differential diagnoses, if targeted or panel testing was negative and for couples who have a family history of a genetic disorder..

-
WES Diagnostic Couple
- It is also performed on the parents of an affected individual or a couple with a family history of a known condition to identify and report any variants relating to the phenotype in question. In addition, a screening test is offered to help identify common recessive variants between couples to attempt and prevent any further genetic disorders. This WES is recommended for affected individuals who have a suspected genetic diagnosis, to help diagnose affected individuals who have multiple differential diagnoses, if targeted or panel testing was negative and for couples who have a family history of a genetic disorder..
-
WES Diagnostic Trio
- It is a comprehensive genetic test that is offered to the affected individual and their parents in order to identify and report any variants relating to the affected individuals disease/condition, additionally this test can help determine whether the disease-causing variant is inherited. . In addition, a screening test is offered to the couple to help identify common recessive variants between couples to attempt and prevent any further genetic disorders. This WES is recommended for affected individuals who have a suspected genetic diagnosis, to help diagnose affected individuals who have multiple differential diagnoses, if targeted or panel testing was negative and for couples who have a family history of a genetic disorder.
-
WES Planning a healthy family
- WES Screening is also an important genetic test that is recommended before planning a family. This test helps determine whether a couple is at risk of having a child with a genetic disorder. If the couple has one or more variants in common, preventative measures can be taken in order to have a healthy child. Carriers of genetic variants are usually healthy individuals. However, when both parents carry a variant in the same gene, they are at risk of having an affected child. This whole exome test is recommended for consanguineous couples and any couples who would like to rule out the risk of having an affected child.
- WES is used in diagnosing or evaluating a genetic disorder where the results are expected to influence medical management and clinical outcomes of a patient or a family directly or indirectly. With the advent of technology, sequencing has become a routine process in clinical diagnosis. In situations where the clinical presentation is unclear and the condition in question is unkonwn, sequencing and analyzing a small number of genes at a time is costly and time-consuming process. This may further delay the diagnosis, which could have an impact on patient’s quality of life.
- WES is a cost-effective diagnostic solution which permits sequencing data from ~24,000 genes from a simple blood draw. WES examines a wider range of genes and variants, which is especially worthwhile for couples looking to know if they are carriers of common recessive disorders and to diagnose genetic disease.
Igenomix uses an internally validated algorithm for precision panel analysis and interpretation. Genetic test results are classified and reported based on the recommendations of the American College of Medical Genetics and Genomics (ACMG) (Richards et al., 2015). According to the guidelines of the ACMG 2015, a genetic variant is classified as either Pathogenic, Likely pathogenic or Benign, Likely benign; any genetic variant which does not fulfill the criteria of pathogenic or benign is classified as a ‘variant of uncertain significance’.
This test aims to identify the molecular cause of the genetic disease in question. A normal genetic result may significantly reduce, but cannot eliminate, the likelihood that the condition is genetic or that a genetic disorder will develop in the future. If any genetic condition is known in the family and molecular testing was already performed, then the specific gene/chromosome variation(s) present in the family should be disclosed at the time of testing.
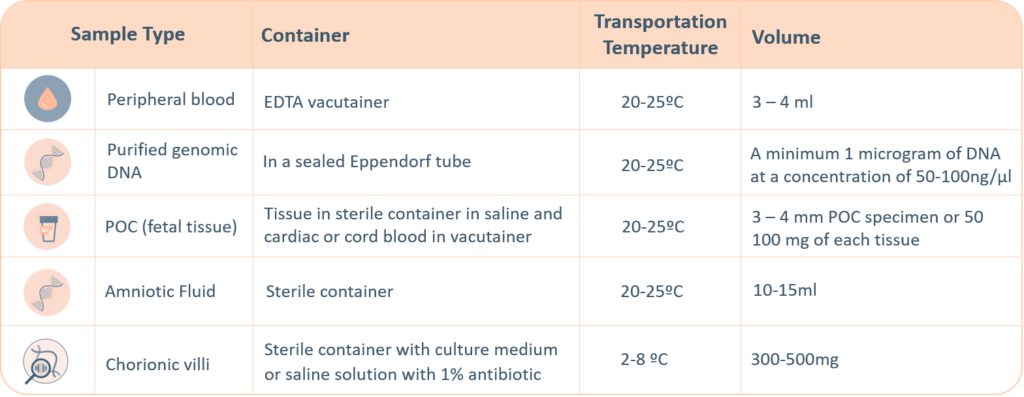
For genetic testing through next generation sequencing, the following sample types are accepted. A thorough labelling of the tube with unique identifying information is suggested, incorrect labelling can lead to rejection of the sample. The minimum required information to identify and accept a sample is – Patient’s full name, Date of birth, Gender and Medical Record Number.
There are four possible results that can be obtained from any precision panel:
-
-
-
Positive (Pathogenic and likely pathogenic):
A positive result indicates that one or more gene or chromosome variation has been identified in association with the disease phenotype. This scenario will allow healthcare professionals to provide genetic counselling or personal guidance regarding possible medical treatments, disease progression, reproductive-/prevention-strategies and potential implications for other family members .
-
-
-
Negative:
A negative result indicates that no disease-causing genetic variant was identified in the test performed. It does not guarantee that the individual will be healthy or free from other genetic disorders or medical conditions. Additionally, a negative result does not rule out a genetic cause of the disease nor does it eliminate the risk for future offspring. However, if a negative test result is obtained and the variant in question is known to be present in affected family members, this then rules out a diagnosis of that genetic disorder in the proband. A negative result may be explained by several causes, including limited genetic knowledge and limitations associated to the used methodology.
-
-
-
Inconclusive/Variant of Uncertain Significance (VUS):
A finding of a variant of uncertain significance indicates that a change in a gene was detected, but it is currently unknown whether that change is associated with a genetic disorder or disease. A variant of uncertain significance is not the same as a positive result and does not clarify whether the proband is at an increased risk of developing a genetic disorder or disease. The change could be a normal genetic variant, or it could be disease-causing. Further analysis may be recommended, including testing both parents as well as other affected and unaffected family members. Sometimes, performing ancillary tests is necessary to prove the phenotype that the proband presents with. Detailed medical records or information from other family members also may be needed to help clarify the result.
-
-
-
Unexpected/Incidental/secondary results:
In rare instances, this test may reveal an important genetic change that is not directly related to the reason for ordering this test. For example, this test may provide information about an individual’s risk for other genetic conditions. This information is likely to impact the individual’s treatment options and is disclosed based on the informed consent provided by the patient.
-
-
Additionally, in accordance to the ACMG guidelines for reporting secondary findings in clinical exome sequencing (PMID: 27854360), pathogenic and likely pathogenic variants in the following genes are reported if consent is indicated on the Test Request Form: ACTA2, ACTC1, APC, APOB, ATP7B, BMPR1A, BRCA1, BRCA2, CACNA1S, COL3A1, DSC2, DSG2, DSP, FBN1, GLA, KCNH2, KCNQ1, LDLR, LMNA, MEN1, MLH1, MSH2, MSH6, MUTYH, MYBPC3, MYH11, MYH7, MYL2, MYL3, NF2, OTC, PCSK9, PKP2, PMS2, PRKAG2, PTEN, RB1, RET, RYR1, RYR2, SCN5A, SDHAF2, SDHB, SDHC, SDHD, SMAD3, SMAD4, STK11, TGFBR1, TGFBR2, TMEM43, TNNI3, TNNT2, TP53, TPM1, TSC1, TSC2, VHL, and WT1. It is encouraged to further ascertain the genotype-phenotype correlation and research to establish the efficacy of intervention in asymptomatic patients with a reported variant in any of the associated genes. This information is only applicable for the whole exome sequencing test and consent must be provided by the patient to obtain this information.
Result interpretation is based on currently available information in the medical literature, research, and scientific databases. Because the literature, medical and scientific knowledge are constantly changing, new information that becomes available in the future may replace or add to the information that Igenomix used to interpret the results. Re-analysis of variants in previously issued reports considering new evidence is not routinely performed but is available upon request.
Sample Requirements
For genetic testing through next generation sequencing, the following sample types are accepted. A thorough labelling of the tube with unique identifying information is suggested, incorrect labelling can lead to rejection of the sample. The minimum required information to identify and accept a sample is – Patient’s full name, Date of birth, Gender and Medical Record Number.
-
-
- Maternal blood sample must be sent with all products of conception, CVS and Amnio samples.
- Precedence will be given to all prenatal samples.
-
The ‘informed consent’ form and the ‘test requisition from’ (included within the provided kit) must be properly filled-in and signed by the patient and sent with the samples inside the shipping box or by e-mail to the laboratory. Igenomix will send you all the documents needed for the pick-up and transportation of the appropriate kit to our laboratory
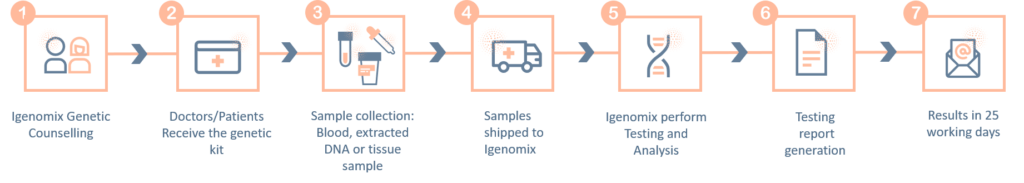
Methodology
Limitations
The probes used for this test are designed to detect known genes in the curated panel. Therefore, this test is unable to detect genes not defined by the NCBI reference genome GRCh37 or non-human genome sequences including viral sequences or non-nuclear DNA that are designated in the specific panel.
In addition, due to the limitations of NGS technologies, the following variants cannot be readily detected: large deletions/duplications greater than 40 base pairs, copy number variations, homopolymer stretches, variants in pseudogene regions, gene fusions, balanced translocations, inversions, ploidy changes, uniparental disomy, and repeat expansion regions.
Furthermore, variants present outside the exons (non-coding region) could be missed; these variants can affect gene activity and protein production which may lead to genetic disorders. This technique does not cover the entire exome, (the % of bases with coverage above 20x is approximately 97%). It may not be possible to resolve certain details about variants such as mosaicism, phasing, or mapping ambiguity.
Analytical limitations may also occur due to the provided clinician information. Accurate and thorough clinical information of the patient(s) and family members is required as incomplete information may lead to false positive or negative results.





